DNA can be used for more than just biology and genetics, but also for applications in fields such as information storage, object marking and traceability.
The extraordinary information density of DNA that can be used for storing data is particularly useful given the explosion of digital data.
Starting from a basic understanding of DNA and its components, let’s take a look at how digital files can be encoded in DNA using the correspondences between A, C, T, and G bases. The processes for writing and reading DNA still have some limitations particularly in synthesizing long strands of DNA, but the revolutionary potential is there.
DNA: Memory resisting the passage of time.
The oldest DNA in the world is 2 million years old. Found in sediments in Greenland, it revealed hundreds of plant species and mastodons which populated Greenland at a time when it was still covered with forest.
This discovery follows on from the work of Svante Pääbo, an interpreter of ancient DNA and winner of the 2022 Nobel Prize for Physiology or Medicine, for his research work on evolutionary genetics and particularly sequencing the Neanderthal nuclear genome. DNA is infinitely small, yet extremely revealing and durable, with possible use beyond palaeontology, biology, genetics and human health.
Researchers in synthetic biology exploit the ability to read, edit and write DNA for products such as vaccines and drug delivery devices or genome engineering to generate organisms with specific properties. But applications for cryptography, information storage, object marking, and traceability are fast-moving innovation areas.
DNA structure.
DNA is made up of four nucleobases (or bases), A, C, T, G, which are connected as base pairs A-T and G-C. Oligonucleotides are short single or double-stranded DNA sequences that can be synthesized in single or double-stranded molecules. Plasmids are small circular DNA fragments found in bacterial and yeast cells that can be synthesized in the lab.
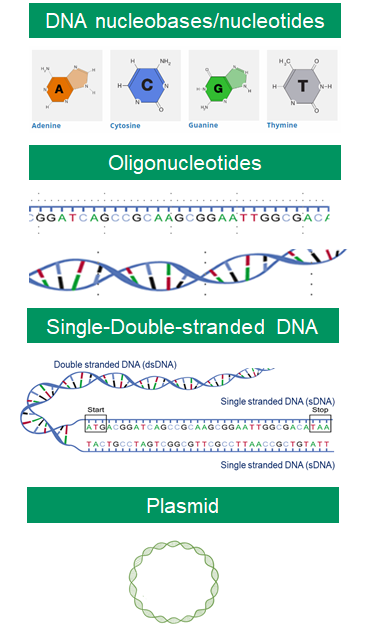
DNA alphabets.
When DNA is read in the cells, it produces proteins for the functioning and reproduction of living beings. Over 3 billion years of evolution, the cellular machinery has developed spellcheckers, word removers and copy-and-paste mechanisms for DNA. Today, synthetic biology allows the same thing to be done and any digital file can be encoded in the form of DNA: text, image, film, PDF, Excel, ZIP …
To convert data from binary format (with 0 and 1) to DNA format (with A, C, T and G bases), the two main options are :
|
1 bit/base : 00 = A , 01 = C , 10 = T , 11 = G 2bits/base : 0 = A or C , 1 = T or G |
Using the above codes, the word “hello” is converted in the corresponding DNA sequences :
DNA says “hello”
| h | e | l | l | o | |
| Binary | 01101000 | 01100101 | 01101100 | 01101100 | 1101111 |
| 2 bits/base | CTTA | CTCC | CTGA | CTGA | CTGG |
| 1 bit/base | AGTCGAAC | ATGCATCG | ATGCTGCA | ATGCTGCA | CTGATGGT |
Writing (or synthetizing) DNA is like telegraphy, where the transmitter made holes in a ribbon to code characters. Writing DNA involves attaching the letters A, C, T, G to each other, through base-to-base addition.
The length of synthesized DNA strands is limited, with oligonucleotide lengths of more than 200 bases (i.e. 200 ACTG bases) representing the current limit of “industrial” production.
Reading technologies are evolving very quickly and allow DNA sequences to be read base by base. One of the most recent techniques uses so called “nanopores”: The oligonucleotides pass through holes. When a base passes through, an electric current is measured. The current is specific to each base, which makes it possible to detect the sequence of the oligonucleotides … and therefore the text. As each fragment has an identifier, the original data can be reassembled in the right order. This technique has the potential to greatly speed up the process of DNA sequencing, making it faster and cheaper than current methods which are still time consuming and expensive.
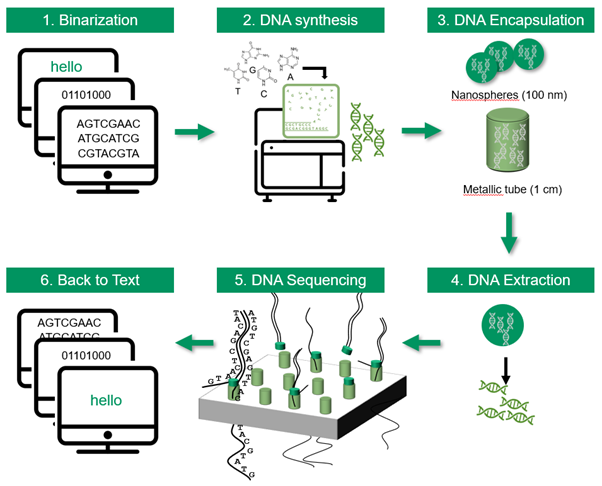
DNA for the dataverse.
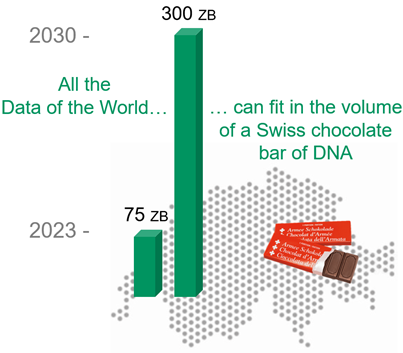
Data storage is a massively expanding domain, with an estimated 75 zettabytes (ZB) (75 1021 bytes) of data to be stored in 2023, 95% of which will never be read again.
It is estimated that in 2030 nearly 300 ZB of data will be stored and that in 2040 one thousandth of the earth’s surface will be used for data centres – 10% of the surface of Switzerland! – to store all these data…
It would take more than 100 million years to download such data with a fibre with a speed of 100 megabits per second, and a serious energy bill!
Data centres alone consume almost 150 TWh per year (150 1012 Wh per year), which equals to 2% of the world’s electricity consumption.
Storing data in DNA is a promising solution since it has an extraordinary information density. All the world’s digital data (i.e. hundreds of ZB) would weigh between 50 and 100 grams of DNA, roughly the weight of a Swiss chocolate bar.
DNA for storage and tracing.
Resistant under specific conditions, DNA remains a fragile molecule, sensitive to high temperatures, humidity, chemical compounds and ultraviolet radiation. To maintain its integrity, it must be protected from these aggressive agents. One way to do this is to encapsulate it for instance in silica microbeads before use. Thus protected, the DNA can be stored or inserted into objects for later reading.
Objects could therefore carry their own immutable memory. A DNA of Things (DoT) for everything and for ever.
However, for DNA to be used as storage, it not only has to be easily written and read, but also easily searchable or indexable in a molecular filing system. This is still at research level, but it has been proven that silica particles used to encapsulate data encoded DNA sequences can also be barcoded or tagged for standard Boolean search. This would complete the absolute potential of DNA as a data carrier other than for biology, but for information storage and traceability.
As with any promising technological development, the issues of privacy and security must be considered so that DNA encoded information does not get misused by unauthorised parties. This calls for a multidisciplinary approach to creating and using synthetic DNA. Just like paleogenomics ermerged as a new discipline following Pääbo’s research, new fields of research and new applications for gene synthesis are already on the way, powered by the need for cost and energy efficient, forever accessible data storage solutions.
